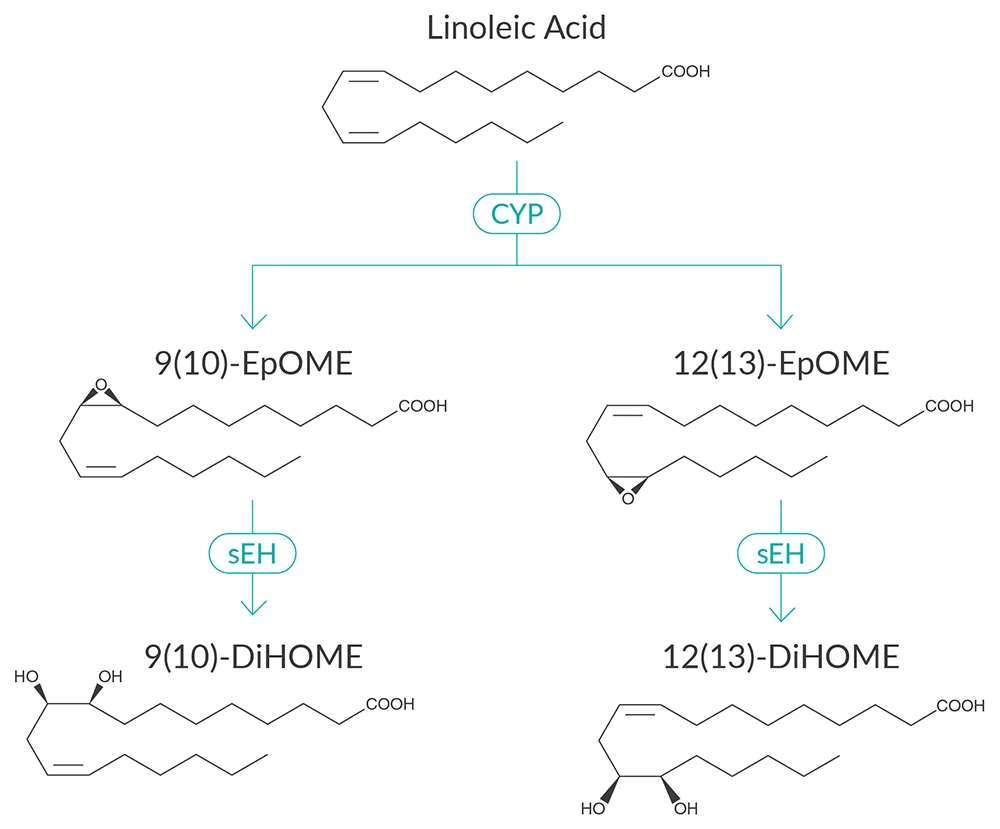
Various fatty acyl lipid mediators derived from dietary polyunsaturated fatty acids (PUFAs) modulate nociception. Many of these lipid mediators can directly bind ion channels, including the Transient Receptor Potential Vanilloid 1 (TRPV1) channel located on sensory nerves. Linoleic acid, known as the most abundant PUFA in modern diet, is a precursor of many pronociceptive lipid mediators-potential TRPV1 binding molecules. Molecular dynamic simulations offers a powerful strategy for interrogating lipid mediator-TRPV1 interactions. The all-atom simulations of TRPV1 channel can be prepared using CHARMM-GUI Membrane Builder. OPM protocols are then used to position and orient the proteins in a lipid membrane bilayer. The systems is relaxed using a sequence of equilibration steps, with the temperature set to 310.15 K using Nose–Hoover coupling, and the pressure set to 1.0 bar using semi-isotropic Parinello–Rahman coupling.The system used in our simulations contains TIP3 water molecules, potassium ions, chlorine ions and lipid molecules. In order to ensure proper system relaxation, two minimization steps and four equilibration steps are used.The lipid composition in the membrane bilayer in all-atom system is fallowing: phosphatidylcholine (POPC), phosphatidylethanolamine (POPE), phosphatidic acid (POPA), phosphatidylserine (POPS), sphingomyelin (PSM), phosphatidylinositol (POPI), cholesterol (CHOL). Each of the molecular dynamic simulations are analyzed by comparing lipid meditor bound and capsaicin-bound structures. Taken together, molecular dynamic workflow can help elucidate molecular mechanism underlying pain modulation by lipid mediators. Although recommendations about fatty acid intake exist for some diseases (e.g. cardiovascular disease), the role of dietary fatty acids in promoting pain disorders is not completely understood.